12. Xarray#
12.1. Overview#
Xarray is a powerful Python library designed for working with multi-dimensional labeled datasets, often used in fields such as climate science, oceanography, and remote sensing. It provides a high-level interface for manipulating and analyzing datasets that can be thought of as extensions of NumPy arrays. Xarray is particularly useful for geospatial data because it supports labeled axes (dimensions), coordinates, and metadata, making it easier to work with datasets that vary across time, space, and other dimensions.
12.2. Learning Objectives#
By the end of this lecture, you should be able to:
Understand the basic concepts and data structures in Xarray, including
DataArrayandDataset.Load and inspect multi-dimensional geospatial datasets using Xarray.
Perform basic operations on Xarray objects, such as selection, indexing, and arithmetic operations.
Use Xarray to efficiently work with large geospatial datasets, including time series and raster data.
Apply Xarray to common geospatial analysis tasks, such as calculating statistics, regridding, and visualization.
12.3. What is Xarray?#
Xarray extends the capabilities of NumPy by providing a data structure for labeled, multi-dimensional arrays. The two main data structures in Xarray are:
DataArray: A labeled, multi-dimensional array, which includes dimensions, coordinates, and attributes.
Dataset: A collection of
DataArrayobjects that share the same dimensions.
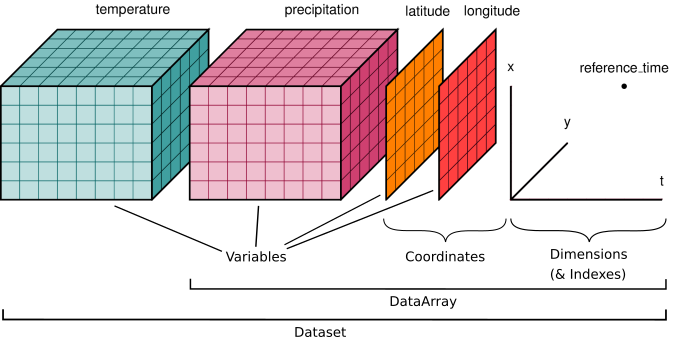
Xarray is particularly useful for working with datasets where dimensions have meaningful labels (e.g., time, latitude, longitude) and where metadata is important.
12.4. Installing Xarray#
Before we start, ensure that Xarray is installed. You can install it via pip:
# %pip install xarray pooch
12.5. Importing Libraries#
import matplotlib.pyplot as plt
import numpy as np
import xarray as xr
xr.set_options(keep_attrs=True, display_expand_data=False)
np.set_printoptions(threshold=10, edgeitems=2)
12.6. Xarray Data Structures#
Xarray provides two core data structures:
DataArray: A single multi-dimensional array with labeled dimensions, coordinates, and metadata.
Dataset: A collection of
DataArrayobjects, each corresponding to a variable, sharing the same dimensions and coordinates.
12.7. Loading a Dataset#
Xarray offers built-in access to several tutorial datasets, which we can load with xr.tutorial.open_dataset. Here, we load an air temperature dataset:
ds = xr.tutorial.open_dataset("air_temperature")
ds
<xarray.Dataset> Size: 31MB
Dimensions: (lat: 25, time: 2920, lon: 53)
Coordinates:
* lat (lat) float32 100B 75.0 72.5 70.0 67.5 65.0 ... 22.5 20.0 17.5 15.0
* lon (lon) float32 212B 200.0 202.5 205.0 207.5 ... 325.0 327.5 330.0
* time (time) datetime64[ns] 23kB 2013-01-01 ... 2014-12-31T18:00:00
Data variables:
air (time, lat, lon) float64 31MB ...
Attributes:
Conventions: COARDS
title: 4x daily NMC reanalysis (1948)
description: Data is from NMC initialized reanalysis\n(4x/day). These a...
platform: Model
references: http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanaly...This dataset is stored in the netCDF format, a common format for scientific data. Xarray automatically parses metadata like dimensions and coordinates.
The dataset is downloaded from the internet and stored in a temporary cache directory. You can find the location of the cache directory depending on your operating system:
Linux:
~/.cache/xarray_tutorial_datamacOS:
~/Library/Caches/xarray_tutorial_dataWindows:
~/AppData/Local/xarray_tutorial_data
12.8. Working with DataArrays#
The DataArray is the core data structure in Xarray. It includes data values, dimensions (e.g., time, latitude, longitude), and the coordinates for each dimension.
# Access a specific DataArray
temperature = ds["air"]
temperature
<xarray.DataArray 'air' (time: 2920, lat: 25, lon: 53)> Size: 31MB
[3869000 values with dtype=float64]
Coordinates:
* lat (lat) float32 100B 75.0 72.5 70.0 67.5 65.0 ... 22.5 20.0 17.5 15.0
* lon (lon) float32 212B 200.0 202.5 205.0 207.5 ... 325.0 327.5 330.0
* time (time) datetime64[ns] 23kB 2013-01-01 ... 2014-12-31T18:00:00
Attributes:
long_name: 4xDaily Air temperature at sigma level 995
units: degK
precision: 2
GRIB_id: 11
GRIB_name: TMP
var_desc: Air temperature
dataset: NMC Reanalysis
level_desc: Surface
statistic: Individual Obs
parent_stat: Other
actual_range: [185.16 322.1 ]You can also access DataArray using dot notation:
ds.air
<xarray.DataArray 'air' (time: 2920, lat: 25, lon: 53)> Size: 31MB
[3869000 values with dtype=float64]
Coordinates:
* lat (lat) float32 100B 75.0 72.5 70.0 67.5 65.0 ... 22.5 20.0 17.5 15.0
* lon (lon) float32 212B 200.0 202.5 205.0 207.5 ... 325.0 327.5 330.0
* time (time) datetime64[ns] 23kB 2013-01-01 ... 2014-12-31T18:00:00
Attributes:
long_name: 4xDaily Air temperature at sigma level 995
units: degK
precision: 2
GRIB_id: 11
GRIB_name: TMP
var_desc: Air temperature
dataset: NMC Reanalysis
level_desc: Surface
statistic: Individual Obs
parent_stat: Other
actual_range: [185.16 322.1 ]12.9. DataArray Components#
Values: The actual data stored in a NumPy array or similar structure.
Dimensions: Named axes of the data (e.g., time, latitude, longitude).
Coordinates: Labels for the values in each dimension (e.g., specific times or geographic locations).
Attributes: Metadata associated with the data (e.g., units, descriptions).
You can extract and print the values, dimensions, and coordinates of a DataArray:
temperature.values
array([[[241.2 , 242.5 , ..., 235.5 , 238.6 ],
[243.8 , 244.5 , ..., 235.3 , 239.3 ],
...,
[295.9 , 296.2 , ..., 295.9 , 295.2 ],
[296.29, 296.79, ..., 296.79, 296.6 ]],
[[242.1 , 242.7 , ..., 233.6 , 235.8 ],
[243.6 , 244.1 , ..., 232.5 , 235.7 ],
...,
[296.2 , 296.7 , ..., 295.5 , 295.1 ],
[296.29, 297.2 , ..., 296.4 , 296.6 ]],
...,
[[245.79, 244.79, ..., 243.99, 244.79],
[249.89, 249.29, ..., 242.49, 244.29],
...,
[296.29, 297.19, ..., 295.09, 294.39],
[297.79, 298.39, ..., 295.49, 295.19]],
[[245.09, 244.29, ..., 241.49, 241.79],
[249.89, 249.29, ..., 240.29, 241.69],
...,
[296.09, 296.89, ..., 295.69, 295.19],
[297.69, 298.09, ..., 296.19, 295.69]]])
temperature.dims
('time', 'lat', 'lon')
temperature.coords
Coordinates:
* lat (lat) float32 100B 75.0 72.5 70.0 67.5 65.0 ... 22.5 20.0 17.5 15.0
* lon (lon) float32 212B 200.0 202.5 205.0 207.5 ... 325.0 327.5 330.0
* time (time) datetime64[ns] 23kB 2013-01-01 ... 2014-12-31T18:00:00
temperature.attrs
{'long_name': '4xDaily Air temperature at sigma level 995',
'units': 'degK',
'precision': np.int16(2),
'GRIB_id': np.int16(11),
'GRIB_name': 'TMP',
'var_desc': 'Air temperature',
'dataset': 'NMC Reanalysis',
'level_desc': 'Surface',
'statistic': 'Individual Obs',
'parent_stat': 'Other',
'actual_range': array([185.16, 322.1 ], dtype=float32)}
12.10. Indexing and Selecting Data#
Xarray allows you to easily select data based on dimension labels, which is very intuitive when working with geospatial data.
# Select data for a specific time and location
selected_data = temperature.sel(time="2013-01-01", lat=40.0, lon=260.0)
selected_data
<xarray.DataArray 'air' (time: 4)> Size: 32B
array([265.2, 266.2, 262.4, 267.5])
Coordinates:
lat float32 4B 40.0
lon float32 4B 260.0
* time (time) datetime64[ns] 32B 2013-01-01 ... 2013-01-01T18:00:00
Attributes:
long_name: 4xDaily Air temperature at sigma level 995
units: degK
precision: 2
GRIB_id: 11
GRIB_name: TMP
var_desc: Air temperature
dataset: NMC Reanalysis
level_desc: Surface
statistic: Individual Obs
parent_stat: Other
actual_range: [185.16 322.1 ]# Slice data across a range of times
time_slice = temperature.sel(time=slice("2013-01-01", "2013-01-31"))
time_slice
<xarray.DataArray 'air' (time: 124, lat: 25, lon: 53)> Size: 1MB
array([[[241.2 , 242.5 , ..., 235.5 , 238.6 ],
[243.8 , 244.5 , ..., 235.3 , 239.3 ],
...,
[295.9 , 296.2 , ..., 295.9 , 295.2 ],
[296.29, 296.79, ..., 296.79, 296.6 ]],
[[242.1 , 242.7 , ..., 233.6 , 235.8 ],
[243.6 , 244.1 , ..., 232.5 , 235.7 ],
...,
[296.2 , 296.7 , ..., 295.5 , 295.1 ],
[296.29, 297.2 , ..., 296.4 , 296.6 ]],
...,
[[238. , 237.7 , ..., 240.89, 242.3 ],
[238.1 , 237.1 , ..., 236.5 , 239. ],
...,
[296.6 , 296.9 , ..., 296.1 , 295.9 ],
[297.5 , 297.4 , ..., 296.79, 296.7 ]],
[[238.1 , 238.39, ..., 240.2 , 241.3 ],
[240.3 , 239.7 , ..., 236.3 , 238.39],
...,
[296.29, 296.6 , ..., 296.5 , 295.9 ],
[297.4 , 297.4 , ..., 297.2 , 296.79]]])
Coordinates:
* lat (lat) float32 100B 75.0 72.5 70.0 67.5 65.0 ... 22.5 20.0 17.5 15.0
* lon (lon) float32 212B 200.0 202.5 205.0 207.5 ... 325.0 327.5 330.0
* time (time) datetime64[ns] 992B 2013-01-01 ... 2013-01-31T18:00:00
Attributes:
long_name: 4xDaily Air temperature at sigma level 995
units: degK
precision: 2
GRIB_id: 11
GRIB_name: TMP
var_desc: Air temperature
dataset: NMC Reanalysis
level_desc: Surface
statistic: Individual Obs
parent_stat: Other
actual_range: [185.16 322.1 ]12.11. Performing Operations on DataArrays#
You can perform arithmetic operations directly on DataArray objects, similar to how you would with NumPy arrays. Xarray also handles broadcasting automatically.
# Calculate the mean temperature over time
mean_temperature = temperature.mean(dim="time")
mean_temperature
<xarray.DataArray 'air' (lat: 25, lon: 53)> Size: 11kB
260.4 260.2 259.9 259.5 259.0 258.6 ... 298.0 297.9 297.8 297.3 297.3 297.3
Coordinates:
* lat (lat) float32 100B 75.0 72.5 70.0 67.5 65.0 ... 22.5 20.0 17.5 15.0
* lon (lon) float32 212B 200.0 202.5 205.0 207.5 ... 325.0 327.5 330.0
Attributes:
long_name: 4xDaily Air temperature at sigma level 995
units: degK
precision: 2
GRIB_id: 11
GRIB_name: TMP
var_desc: Air temperature
dataset: NMC Reanalysis
level_desc: Surface
statistic: Individual Obs
parent_stat: Other
actual_range: [185.16 322.1 ]# Subtract the mean temperature from the original data
anomalies = temperature - mean_temperature
anomalies
<xarray.DataArray 'air' (time: 2920, lat: 25, lon: 53)> Size: 31MB
-19.18 -17.68 -16.39 -15.48 -14.92 ... -0.5012 -0.6276 -0.8482 -1.091 -1.615
Coordinates:
* lat (lat) float32 100B 75.0 72.5 70.0 67.5 65.0 ... 22.5 20.0 17.5 15.0
* lon (lon) float32 212B 200.0 202.5 205.0 207.5 ... 325.0 327.5 330.0
* time (time) datetime64[ns] 23kB 2013-01-01 ... 2014-12-31T18:00:00
Attributes:
long_name: 4xDaily Air temperature at sigma level 995
units: degK
precision: 2
GRIB_id: 11
GRIB_name: TMP
var_desc: Air temperature
dataset: NMC Reanalysis
level_desc: Surface
statistic: Individual Obs
parent_stat: Other
actual_range: [185.16 322.1 ]12.12. Visualization with Xarray#
Xarray integrates well with Matplotlib and other visualization libraries, making it easy to create plots directly from DataArray and Dataset objects.
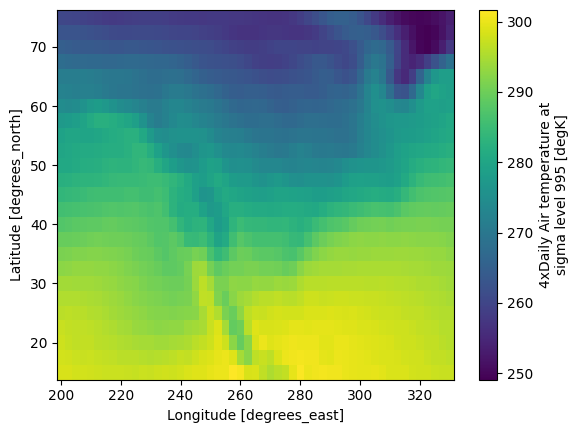
# Plot the mean temperature
mean_temperature.plot()
plt.show()
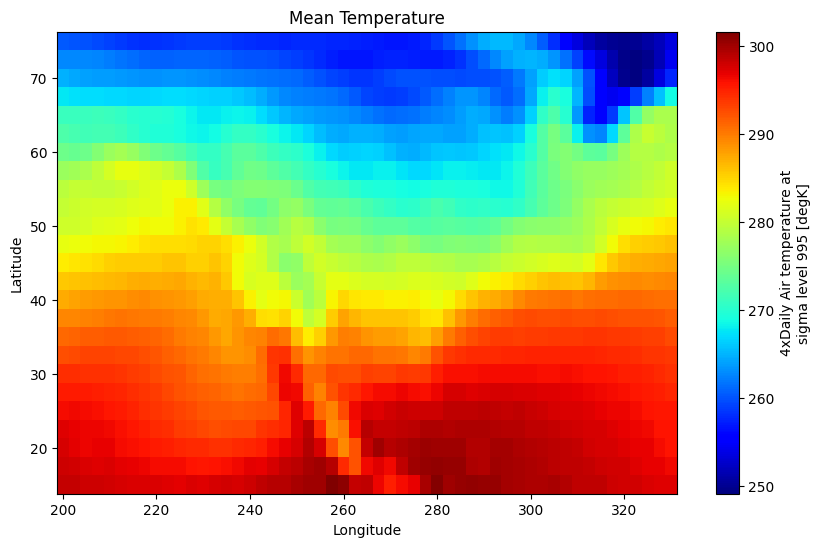
You can customize the appearance of plots by passing arguments to the plot method. For example, you can specify the color map, add labels, and set the figure size.
mean_temperature.plot(cmap="jet", figsize=(10, 6))
plt.xlabel("Longitude")
plt.ylabel("Latitude")
plt.title("Mean Temperature")
Text(0.5, 1.0, 'Mean Temperature')
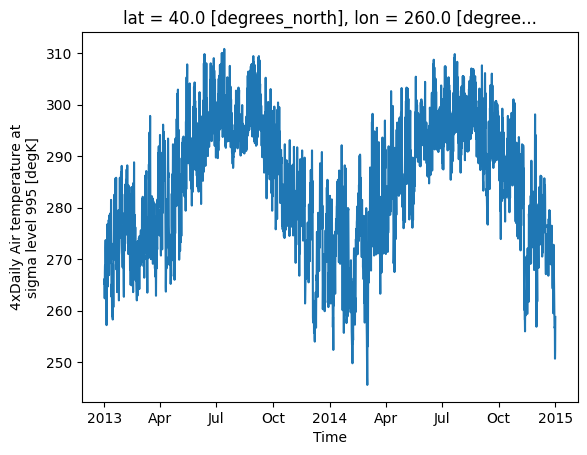
You can also select a specific location using the sel method and plot a time series of temperature at that location.
# Plot a time series for a specific location
temperature.sel(lat=40.0, lon=260.0).plot()
plt.show()
12.13. Working with Datasets#
A Dataset is a collection of DataArray objects. It is useful when you need to work with multiple related variables.
# List all variables in the dataset
print(ds.data_vars)
Data variables:
air (time, lat, lon) float64 31MB 241.2 242.5 243.5 ... 296.2 295.7
# Access a DataArray from the Dataset
temperature = ds["air"]
# Perform operations on the Dataset
mean_temp_ds = ds.mean(dim="time")
mean_temp_ds
<xarray.Dataset> Size: 11kB
Dimensions: (lat: 25, lon: 53)
Coordinates:
* lat (lat) float32 100B 75.0 72.5 70.0 67.5 65.0 ... 22.5 20.0 17.5 15.0
* lon (lon) float32 212B 200.0 202.5 205.0 207.5 ... 325.0 327.5 330.0
Data variables:
air (lat, lon) float64 11kB 260.4 260.2 259.9 ... 297.3 297.3 297.3
Attributes:
Conventions: COARDS
title: 4x daily NMC reanalysis (1948)
description: Data is from NMC initialized reanalysis\n(4x/day). These a...
platform: Model
references: http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanaly...12.14. Why Use Xarray?#
Xarray is valuable for handling multi-dimensional data, especially in scientific applications. It provides metadata, dimension names, and coordinate labels, making it much easier to understand and manipulate data compared to raw NumPy arrays.
12.14.1. Without Xarray (Using NumPy)#
Here’s how a task might look without Xarray, using NumPy arrays:
lat = ds.air.lat.data
lon = ds.air.lon.data
temp = ds.air.data
temp.shape
(2920, 25, 53)
plt.figure()
plt.pcolormesh(lon, lat, temp[0, :, :])
<matplotlib.collections.QuadMesh at 0x7f29acf11670>
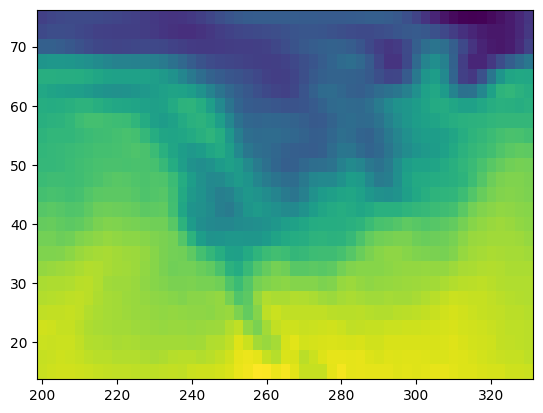
While this approach works, it’s not clear what 0 refers to (in this case, it’s the first time step).
12.14.2. With Xarray#
With Xarray, you can use more intuitive and readable indexing with sel and isel:
ds.air.isel(time=0).plot(x="lon")
<matplotlib.collections.QuadMesh at 0x7f29acf126f0>
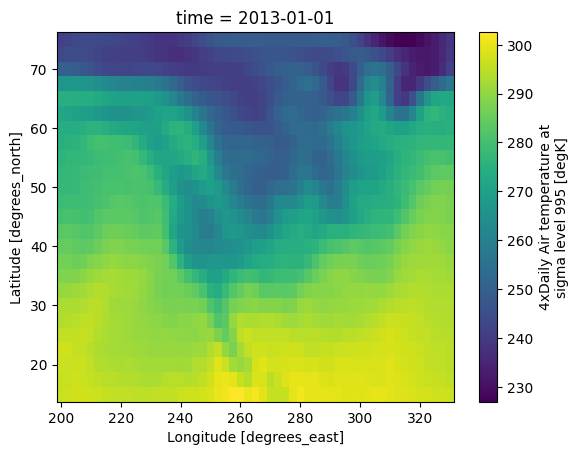
ds.air.sel(time="2013-01-01T00:00:00").plot(x="lon")
<matplotlib.collections.QuadMesh at 0x7f29acf11790>

This example selects the first time step and plots it using labeled axes (lat and lon), which is much clearer.
12.15. Advanced Indexing: Label vs. Position-Based Indexing#
Xarray supports both label-based and position-based indexing, making it flexible for data selection.
12.15.1. Label-based Indexing#
You can use .sel() to select data based on the labels of coordinates, such as specific times or locations:
# Select all data from May 2013
ds.sel(time="2013-05")
<xarray.Dataset> Size: 1MB
Dimensions: (lat: 25, time: 124, lon: 53)
Coordinates:
* lat (lat) float32 100B 75.0 72.5 70.0 67.5 65.0 ... 22.5 20.0 17.5 15.0
* lon (lon) float32 212B 200.0 202.5 205.0 207.5 ... 325.0 327.5 330.0
* time (time) datetime64[ns] 992B 2013-05-01 ... 2013-05-31T18:00:00
Data variables:
air (time, lat, lon) float64 1MB 259.2 259.3 259.1 ... 297.6 297.5
Attributes:
Conventions: COARDS
title: 4x daily NMC reanalysis (1948)
description: Data is from NMC initialized reanalysis\n(4x/day). These a...
platform: Model
references: http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanaly...# Slice over time, selecting data between May and July 2013
ds.sel(time=slice("2013-05", "2013-07"))
<xarray.Dataset> Size: 4MB
Dimensions: (lat: 25, time: 368, lon: 53)
Coordinates:
* lat (lat) float32 100B 75.0 72.5 70.0 67.5 65.0 ... 22.5 20.0 17.5 15.0
* lon (lon) float32 212B 200.0 202.5 205.0 207.5 ... 325.0 327.5 330.0
* time (time) datetime64[ns] 3kB 2013-05-01 ... 2013-07-31T18:00:00
Data variables:
air (time, lat, lon) float64 4MB 259.2 259.3 259.1 ... 299.5 299.7
Attributes:
Conventions: COARDS
title: 4x daily NMC reanalysis (1948)
description: Data is from NMC initialized reanalysis\n(4x/day). These a...
platform: Model
references: http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanaly...12.15.2. Position-based Indexing#
Alternatively, you can use .isel() to select data based on the positions of coordinates:
# Select the first time step, second latitude, and third longitude
ds.air.isel(time=0, lat=2, lon=3)
<xarray.DataArray 'air' ()> Size: 8B
array(247.5)
Coordinates:
lat float32 4B 70.0
lon float32 4B 207.5
time datetime64[ns] 8B 2013-01-01
Attributes:
long_name: 4xDaily Air temperature at sigma level 995
units: degK
precision: 2
GRIB_id: 11
GRIB_name: TMP
var_desc: Air temperature
dataset: NMC Reanalysis
level_desc: Surface
statistic: Individual Obs
parent_stat: Other
actual_range: [185.16 322.1 ]12.16. High-Level Computations with Xarray#
Xarray offers several high-level operations that make common computations straightforward, such as groupby, resample, rolling, and weighted.
12.16.1. GroupBy Operation#
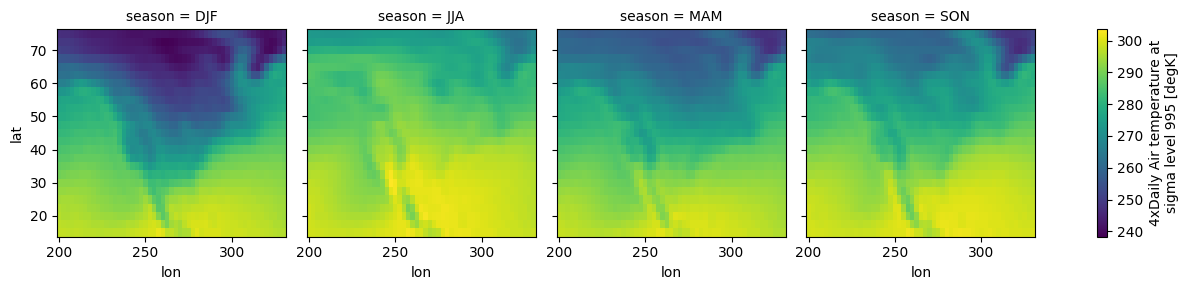
You can calculate statistics such as the seasonal mean of the dataset:
seasonal_mean = ds.groupby("time.season").mean()
seasonal_mean.air.plot(col="season")
<xarray.plot.facetgrid.FacetGrid at 0x7f29acd4ff20>
12.16.2. Computation with Weights#
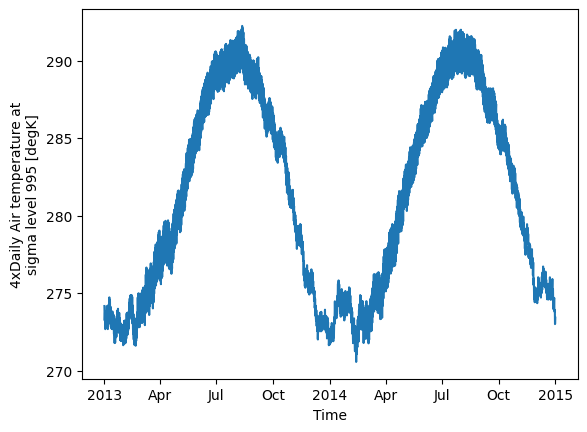
Xarray allows for weighted computations, useful in geospatial contexts where grid cells vary in size. For example, you can weight the mean of the dataset by cell area.
cell_area = xr.ones_like(ds.air.lon) # Placeholder for actual area calculation
weighted_mean = ds.weighted(cell_area).mean(dim=["lon", "lat"])
weighted_mean.air.plot()
[<matplotlib.lines.Line2D at 0x7f29acd26300>]
12.16.3. Rolling Window Operation#
Xarray supports rolling window operations, which are useful for smoothing time series data spatially or temporally. For example, you can smooth the temperature data spatially using a 5x5 window.
ds.air.isel(time=0).rolling(lat=5, lon=5).mean().plot()
<matplotlib.collections.QuadMesh at 0x7f29acb57a70>
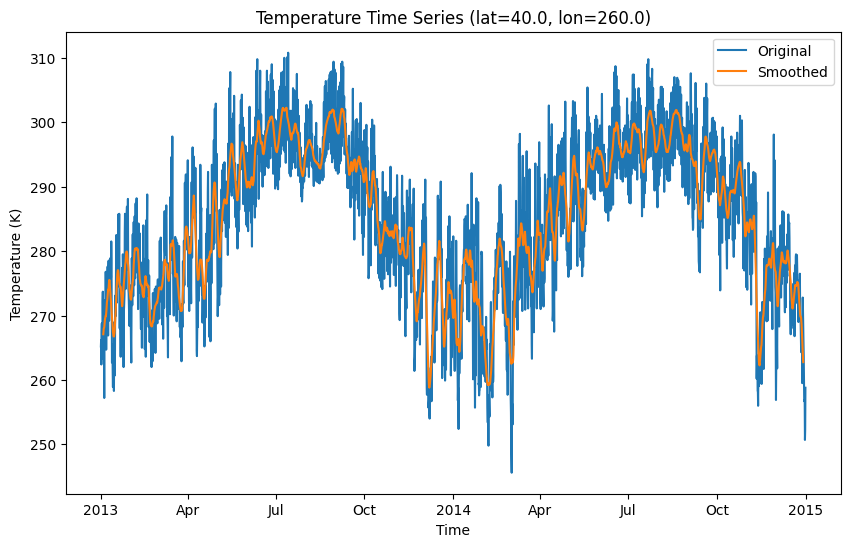
Similarly, you can smooth the temperature data temporally using a 5-day window.
plt.figure(figsize=(10, 6))
# Select the time series at a specific latitude and longitude
temperature = ds["air"].sel(lat=40.0, lon=260.0)
# Plot the original time series
temperature.plot(label="Original")
# Apply rolling mean smoothing with a window size of 20
smoothed_temperature = temperature.rolling(time=20, center=True).mean()
# Plot the smoothed data
smoothed_temperature.plot(label="Smoothed")
# Add a title and labels
plt.title("Temperature Time Series (lat=40.0, lon=260.0)")
plt.xlabel("Time")
plt.ylabel("Temperature (K)")
# Add a legend
plt.legend()
# Show the plot
plt.show()
12.17. Reading and Writing Files#
Xarray supports many common scientific data formats, including netCDF and Zarr. You can read and write datasets to disk with a few simple commands.
12.17.1. Writing to netCDF#
To save a dataset as a netCDF file:
# Ensure air is in a floating-point format (float32 or float64)
ds["air"] = ds["air"].astype("float32")
# Save the dataset to a NetCDF file
ds.to_netcdf("air_temperature.nc")
12.17.2. Reading from netCDF#
To load a dataset from a netCDF file:
loaded_data = xr.open_dataset("air_temperature.nc")
loaded_data
<xarray.Dataset> Size: 15MB
Dimensions: (lat: 25, time: 2920, lon: 53)
Coordinates:
* lat (lat) float32 100B 75.0 72.5 70.0 67.5 65.0 ... 22.5 20.0 17.5 15.0
* lon (lon) float32 212B 200.0 202.5 205.0 207.5 ... 325.0 327.5 330.0
* time (time) datetime64[ns] 23kB 2013-01-01 ... 2014-12-31T18:00:00
Data variables:
air (time, lat, lon) float32 15MB ...
Attributes:
Conventions: COARDS
title: 4x daily NMC reanalysis (1948)
description: Data is from NMC initialized reanalysis\n(4x/day). These a...
platform: Model
references: http://www.esrl.noaa.gov/psd/data/gridded/data.ncep.reanaly...12.18. Exercises#
12.18.1. Exercise 1: Exploring a New Dataset#
Load the Xarray tutorial dataset
rasm.Inspect the
Datasetobject and list all the variables and dimensions.Select the
Tairvariable (air temperature).Print the attributes, dimensions, and coordinates of
Tair.
12.18.2. Exercise 2: Data Selection and Indexing#
Select a subset of the
Tairdata for the date1980-07-01and latitude70.0.Create a time slice for the entire latitude range between January and March of 1980.
Plot the selected time slice as a line plot.
12.18.3. Exercise 3: Performing Arithmetic Operations#
Compute the mean of the
Tairdata over thetimedimension.Subtract the computed mean from the original
Tairdataset to get the temperature anomalies.Plot the mean temperature and the anomalies on separate plots.
12.18.4. Exercise 4: GroupBy and Resampling#
Use
groupbyto calculate the seasonal mean temperature (Tair).Use
resampleto calculate the monthly mean temperature for 1980.Plot the seasonal mean for each season and the monthly mean.
12.18.5. Exercise 5: Writing Data to netCDF#
Select the temperature anomalies calculated in Exercise 3.
Convert the
Tairvariable tofloat32to optimize file size.Write the anomalies data to a new netCDF file named
tair_anomalies.nc.Load the data back from the file and print its contents.
12.19. Summary#
Xarray is a powerful library for working with multi-dimensional geospatial data. It simplifies data handling by offering labeled dimensions and coordinates, enabling intuitive operations and making analysis more transparent. Xarray’s ability to work seamlessly with NumPy, Dask, and Pandas makes it an essential tool for geospatial and scientific analysis. With Xarray, you can efficiently manage and analyze large, complex datasets, making it a valuable asset for researchers and developers alike.


